Abstract
Since the early 2000s, several event-based surveillance (EBS) systems have been developed to collect information on epidemics published in online media (for example, ProMED-mail since 1994, HealthMap since 2006, PADI-web since 2016, EIOS since 2017). These systems cover a range of diseases and syndromes in both humans and animals. EBS systems serve as alert systems for disease events, enabling epidemiologists to carry out early surveillance, including in zones that are poorly covered by official surveillance systems. Some of these EBS systems, such as PADI-web, integrate artificial intelligence and natural language processing (NLP) to improve automatic document classification and identify epidemiological events within articles. This article will provide examples of the day-to-day use of EBS systems in France.
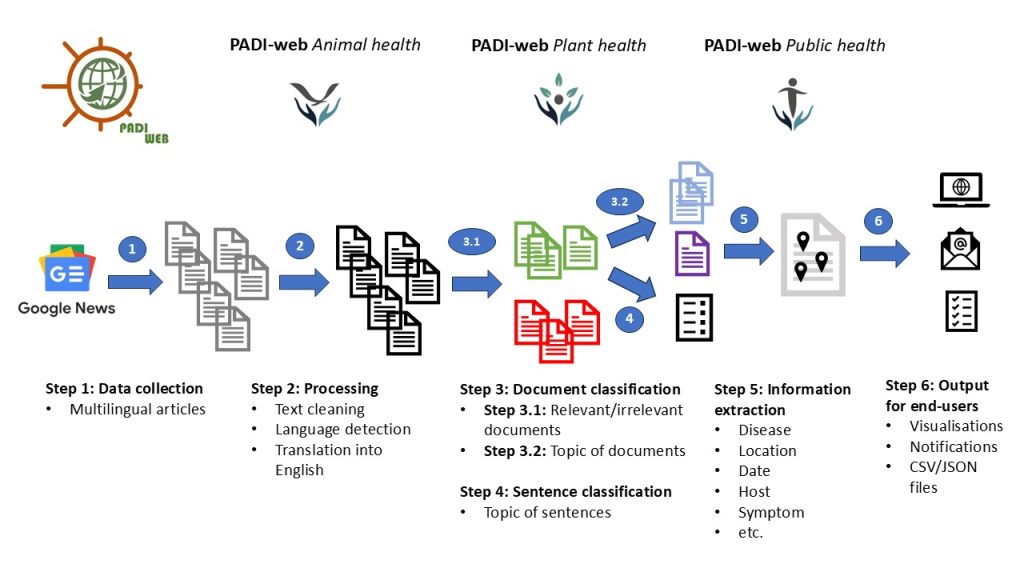
Figure 1. PADI-web processing procedures
Epidemic intelligence is a discipline that is of increasing importance for the early identification of emerging and infectious animal diseases that can endanger the health of a national herd and threaten both public health (zoonotic diseases) and species conservation (species whose numbers are very low, wildlife species). Its aim is to identify, analyse and monitor signals of health hazards that are threatening animal health. It relies on the continuous surveillance of various types of signal from official and non-official sources (media, social media, etc.).
One of the missions of the World Organisation for Animal Health (WOAH) is to improve both the early detection of animal diseases and the dissemination of information on the appearance of these diseases at international level. WOAH members make official information on notifiable animal diseases available, in a structured and standard format, through the database of the World Animal Health Information System (WAHIS). Experts in epidemic intelligence can analyse these data using a set procedure, making it possible to determine epidemiological indicators and carry out periodic situation assessments. However, official data can be subject to bias or notification delays for multiple reasons [1].
A New Approach: Health events extracted from the Web
Since the 2000s, a new generation of health surveillance systems has been developed that complement the existing system by identifying disease events extracted from the internet and other electronic media. These new systems are known as event-based surveillance (EBS) systems [2]. Numerous interfaces have emerged in the public health sector, all of which include an animal health component (e.g. ProMED in 1994, HealthMap in 2006, PADI-web in 2016 and EIOS in 2018). They make it possible to identify, extract, classify and visualise disease events from unstructured textual data.
Some of these EBS systems, such as PADI-web (Platform for Automated extraction of animal Disease Information from the Web) [3,4] integrate artificial intelligence (AI) and natural language processing to improve automatic document classification (see steps 3 and 4 of Figure 1) and identify epidemiological information within articles (see step 5 of Figure 1). Interfaces for information retrieval (Figure 2) and visualisation of epidemiological events are provided (Figure 3).
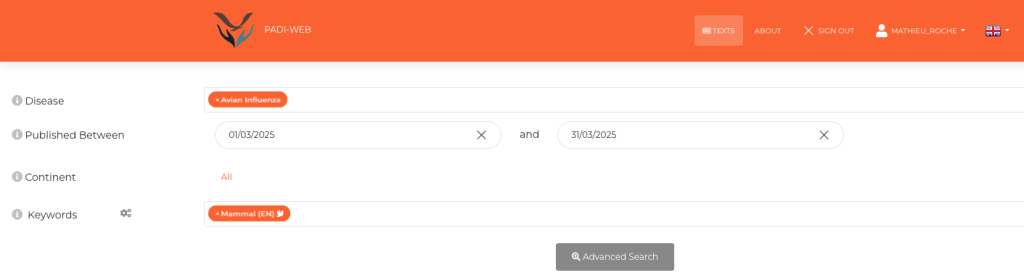
Figure 2. PADI-web search interface
To utilise and adapt AI for animal health surveillance, PADI-web uses data that have been annotated manually by experts. Raw data that have been collected and annotated for animal health purposes are a precious resource that can be employed to train specific models using, for example, machine learning methods. Moreover, AI systems based on general language models (e.g. Bidirectional Encoder Representations from Transformers – BERT [6]) and more specialised models (e.g. BioBERT for the biomedical field [7], AgriBERT for agriculture [8]) enable us to adapt models to specific domains and specific tasks. BERT is a language model with a specific type of architecture that enables it to ‘understand’ the relationships between words in a given sentence.
As part of the MOOD project (MOnitoring Outbreaks for Disease surveillance in a data science context [coordinated by Elena Arsevska]), the French Agricultural Research Centre for International Development (CIRAD) and the French National Research Institute for Agriculture, Food and Environment (INRAE) have carried out multi-disciplinary work to integrate AI models into epidemiological surveillance tools. To build traditional machine learning models (Support Vector Machine, Random Forest, etc.) and/or to adjust language models, dedicated corpora (collections of texts) have been created by arranging data annotation sessions with animal health experts and organising ‘hackathons’ (collaborative events during which people work together on a specific task). These annotation sessions can use inter-annotator agreements and methods such as the Delphi method (several rounds of annotation to reach a consensus) to develop annotation guidelines and improve the quality of the data produced for use by AI algorithms.
The corpora used are annotated at the article level, determining if a text is relevant or not. A text is considered relevant if it describes a new, suspected, or unknown epidemiological outbreak. To develop an information extraction model using relevant texts, the epidemiological information (for example, diseases, hosts, locations, dates, etc.) must also be extracted from the text. AI models that have been trained and adapted through a process of fine-tuning using these annotated data have now been integrated into the PADI-web system.
These AI approaches make it possible to automatically determine (i) whether the documents automatically collected from the web through a process of analysis and indexing are relevant, (ii) what topics they discuss, and (iii) what epidemiological information is contained in the text (diseases, outbreak locations, hosts, symptoms, number of cases, etc.). This integration takes into account the qualitative aspects of the results, but also the simplicity of the algorithms used, which is another important consideration for the AI modules that are currently being developed.
Today, numerous other questions are being studied, for example, how to recognise weak signals or determine identical or new events. In this context, language models and large language models (generative models based on ChatGPT and others) can be particularly effective, as shown by the collaborative work carried out by researchers at CIRAD and Strathmore University (Kenya) [9].
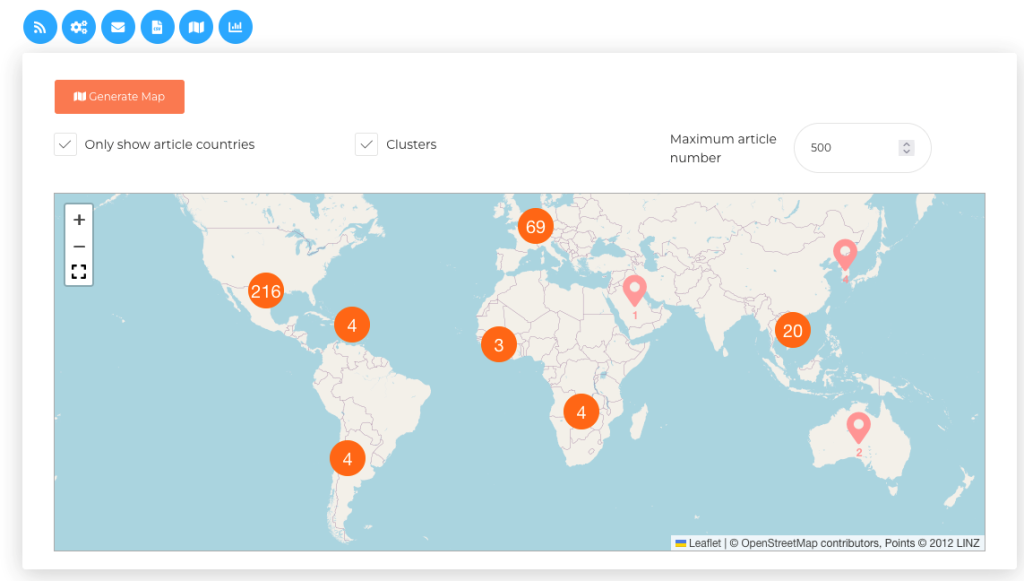
Figure 3. PADI-web interface for spatial data visualisation for epidemiological events
CIRAD and INRAE are also working on syndromic surveillance in animal health, which makes it possible to identify potential new diseases in new locations or with new hosts. As part of this work, a retrospective analysis of the detection of highly pathogenic avian influenza (HPAI) virus in unusual host species was carried out. Seven case studies of HPAI in mammals were identified in the WAHIS database, for which the associated articles collected by PADI-web were validated manually. [10]. Several strategies for classifying locations were evaluated, in order to enhance/optimise the sensitivity and specificity of the surveillance system.
Alongside the research carried out as part of the MOOD project, training sessions for end users have been organised and datasheets [11] have been produced to make it easier to use this tool.
The epidemic intelligence unit of the French National Animal Health Surveillance platform (the ESA platform) [12] uses EBS systems on a daily basis, and one of the challenges is combining all the different epidemiological information from the various different sources. To facilitate this process, a new tool known as MUST (Multi-Source Surveillance Tool) has been developed to establish links between different sources of health data [10]. The first component of MUST focuses on the surveillance of outbreaks of highly pathogenic avian influenza in mammals (HPAI-M).
The tool collects, filters and maps HPAI-M events from official notifications extracted from the WAHIS database, official data from online databases managed by health authorities (USDA for the United States of America and APHA for the United Kingdom), and events from different EBS systems (ProMed-mail and PADI-web). The tool is currently being tested to evaluate different merging strategies based on spatial and temporal criteria, as well as information about the hosts involved in the epidemiological events. This expertise will enable us to put forward approaches for future MUST users.
AI approaches can be particularly effective in addressing animal health surveillance challenges. However, such approaches require expert knowledge to annotate data, validate algorithm outputs, adjust parameters and render the tools operational, all of which calls for multi-disciplinary research.
Translated from the original in French.
Main image copyright: akinbostanci
References
[1] Lin SY, Beltran-Alcrudo D, Awada L, Hamilton-West C, Lavarello Schettini A, Cáceres P, et al. Analysing WAHIS Animal Health Immediate Notifications to Understand Global Reporting Trends and Measure Early Warning Capacities (2005–2021). Transbound. Emerg. Dis. 2023. https://doi.org/10.1155/2023/6666672
[2] Paquet C, Coulombier D, Kaiser R, Ciotti M. Epidemic intelligence: a new framework for strengthening disease surveillance in Europe. Euro Surveill. 2026:11(12);5‑6. https://doi.org/10.2807/esm.11.12.00665-en
[3] Arsevska E, Valentin S, Rabatel J, de Goër de Hervé J, Falala S, Lancelot R, et al. Web monitoring of emerging animal infectious diseases integrated in the French Animal Health Epidemic Intelligence System. PLoS One. 2018:13(8);25. https://doi.org/10.1371/journal.pone.0199960
[4] Valentin S, Arsevska E, Rabatel J, Falala S, Mercier A, Lancelot R, et al. PADI-web 3.0: A new framework for extracting and disseminating fine-grained information from the news for animal disease surveillance. One Health. 2021:13. https://doi.org/10.1016/j.onehlt.2021.100357
[5] Sobkowich KE. Demystifying artificial intelligence for veterinary professionals: practical applications and future potential. Am. J. Vet. Res. 2025:86. https://doi.org/10.2460/ajvr.24.09.0275
[6] Devlin J, Chang MW, Lee K, Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. 2019:1;4171-86. https://doi.org/10.18653/v1/N19-1423
[7] Lee J, Yoon W, Kim S, Kim D, Kim S, Ho So C, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text. Bioinformatics. 2020:36(4);1234-40. https://doi.org/10.1093/bioinformatics/btz682
[8] Rezayi S, Liu Z, Wu Z, Dhakal C, Ge B, Zhen C, et al. AgriBERT: Knowledge-Infused Agricultural Language Models for Matching Food and Nutrition. Proceedings of the Thirty-First International Joint Conference on Artificial Intelligence – AI for Good. 2022;5150-56. https://doi.org/10.24963/ijcai.2022/715
[9] Menya E, Roche M, Interdonato R, Owuor D. EpidGPT: A combined strategy to discriminate between redundant and new information for epidemiological surveillance systems. In: Métais E, Meziane F, Saraee M, Sugumaran V, Valtchev P, eds. Natural Language Processing and Information Systems: 29th International Conference on Applications of Natural Language to Information Systems; 2024 June 25-27, Turin, Italy. Springer-Verlag, Berlin, Heidelberg, p. 439-54. https://doi.org/10.1007/978-3-031-70239-6
[10] Trevennec C, Pompidor P, Bououda S, Rabatel J, Roche M. MUST-AI: Multisource Surveillance Tool – Avian Influenza. Procedia Comput. Sci. 2024:246;3034‑43. https://doi.org/10.1016/j.procs.2024.09.718
[11] Roche M. Data sheets highlighting characteristics of PADI-web. Paris (France): Agritrop CIRAD; 2025. Available at: https://agritrop.cirad.fr/611480/ (accessed on 23 May 2025).
[12] Dupuy C, Locquet C, Brard C, Dommergues L, Faure E, Gache K, et al. The French National Animal Health Surveillance Platform: an innovative, cross-sector collaboration to improve surveillance system efficiency in France and a tangible example of the One Health approach. Front. Vet. Sci. 2024:11. https://doi.org/10.3389/fvets.2024.1249925








